[rev_slider alias=”conifer_embryogenesis”]
Molecular aspects of plant embryo development, begining with a fertilized egg and culminating in a newly germinated plant, have been broadly studied in angiosperm models. However, these aspects have been far less studied in the gymnosperms which include some of the most representative conifer species (e.g. pines) covering a large part of the northern hemisphere landscapes. Given their divergence from a common ancestor more than 300 million years ago, the gymnosperms and the angiosperms present some different charcateristics during embryogenesis that suggest a differential molecular regulation in specific stages of embryo development.
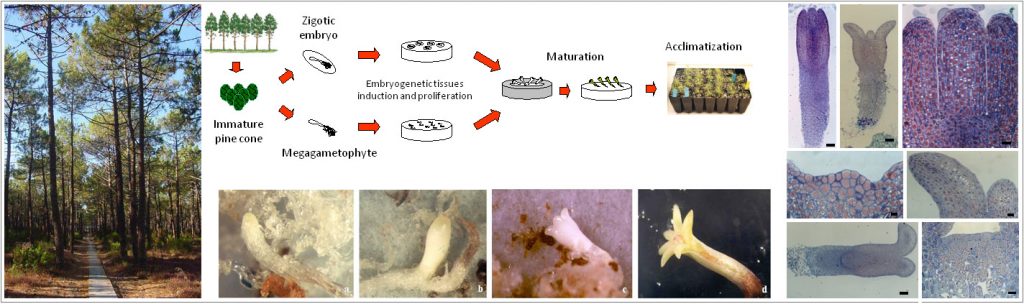
Histological studies on somatic embryogenesis of maritime pine (Pinus pinaster)
We have previously identified a set of transcriptional and post-transcriptional regulators of embryogenesis in maritime pine (Pinus pinaster)(Vega-Bartol et al. 2013; Rodrigues et al. 2018; Rodrigues et al. 2019) and are now conducting functional studies in embryogenic in vitro cultures to characterize the roles of specific microRNAs and their interaction with target genes.
The methodologies being used include transcriptomics and bioinformatic analysis, molecular cloning, plant genetic transformation and somatic embryogenesis (development of embryos from somatic cells withouh the fusion of gametes) in pine and Arabidopsis and Picea abies model systems.
Understanding specific mechanisms underlying the characteristic features of conifer embryogenesis may help to optimize biotechnological applications, such as large-scale clonal propagation of these species via somatic embryogenesis, by assisting in the selection of conditions favoring specific metabolic pathways able to sustain embryo development.
